1.Load packages we will use
Download \(CO_2\) emissions per capita from Our World in Data into the directory for this post.
Assign the location of the file to ‘file.csv’. The data should be in the same directory as this file Read the data into R and assign it to ‘emissions’
file_csv <- here("_posts",
"2021-02-26-reading-and-writing-data",
"co-emissions-per-capita.csv")
emissions <- read_csv(file_csv)
- Show the first 20 rows (observations of) ‘emissions’
emissions
# A tibble: 22,383 x 4
Entity Code Year `Per capita CO2 emissions`
<chr> <chr> <dbl> <dbl>
1 Afghanistan AFG 1949 0.00191
2 Afghanistan AFG 1950 0.0109
3 Afghanistan AFG 1951 0.0117
4 Afghanistan AFG 1952 0.0115
5 Afghanistan AFG 1953 0.0132
6 Afghanistan AFG 1954 0.0130
7 Afghanistan AFG 1955 0.0186
8 Afghanistan AFG 1956 0.0218
9 Afghanistan AFG 1957 0.0343
10 Afghanistan AFG 1958 0.0380
# ... with 22,373 more rows- Start with ‘emissions’ data THEN
use ‘clean_names’ from the janitor package to make the names easier to work with assign the output to ‘tidy_emissions’ show the first 10 rows of ‘tidy_emissions’
tidy_emissions <- emissions %>%
clean_names()
tidy_emissions
# A tibble: 22,383 x 4
entity code year per_capita_co2_emissions
<chr> <chr> <dbl> <dbl>
1 Afghanistan AFG 1949 0.00191
2 Afghanistan AFG 1950 0.0109
3 Afghanistan AFG 1951 0.0117
4 Afghanistan AFG 1952 0.0115
5 Afghanistan AFG 1953 0.0132
6 Afghanistan AFG 1954 0.0130
7 Afghanistan AFG 1955 0.0186
8 Afghanistan AFG 1956 0.0218
9 Afghanistan AFG 1957 0.0343
10 Afghanistan AFG 1958 0.0380
# ... with 22,373 more rows- Start with the ‘tidy_emissions’ THEN use ‘filter’ to extract rows with ‘year == 1985’ THEN use ‘skim’ to calculate the descriptive statistics
| Name | Piped data |
| Number of rows | 209 |
| Number of columns | 4 |
| _______________________ | |
| Column type frequency: | |
| character | 2 |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| entity | 0 | 1.00 | 4 | 32 | 0 | 209 | 0 |
| code | 12 | 0.94 | 3 | 8 | 0 | 197 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| year | 0 | 1 | 1985.00 | 0.00 | 1985.00 | 1985.00 | 1985.00 | 1985.00 | 1985.00 | ▁▁▇▁▁ |
| per_capita_co2_emissions | 0 | 1 | 5.53 | 8.94 | 0.04 | 0.51 | 2.65 | 7.62 | 83.83 | ▇▁▁▁▁ |
- 13 observations have a missing code. How are these observations different? Start with ‘tidy_emissions’ then extract rows with ‘year == 1985’ and are missing a code
# A tibble: 12 x 4
entity code year per_capita_co2_emissions
<chr> <chr> <dbl> <dbl>
1 Africa <NA> 1985 1.23
2 Asia <NA> 1985 1.81
3 Asia (excl. China & India) <NA> 1985 2.76
4 EU-27 <NA> 1985 9.19
5 EU-28 <NA> 1985 9.28
6 Europe <NA> 1985 10.9
7 Europe (excl. EU-27) <NA> 1985 13.3
8 Europe (excl. EU-28) <NA> 1985 14.1
9 North America <NA> 1985 13.2
10 North America (excl. USA) <NA> 1985 5.01
11 Oceania <NA> 1985 10.8
12 South America <NA> 1985 1.87Entities that are not countries do not have country codes.
- Start with tidy_emissions THEN use ‘filter’ to extract rows with year == 1985 without missing codes THEN use ‘select’ to drop the ‘year’ variable THEN use ‘rename’ to change the variable ‘entity’ to ‘country’ assign the output to ‘emissions_1985’
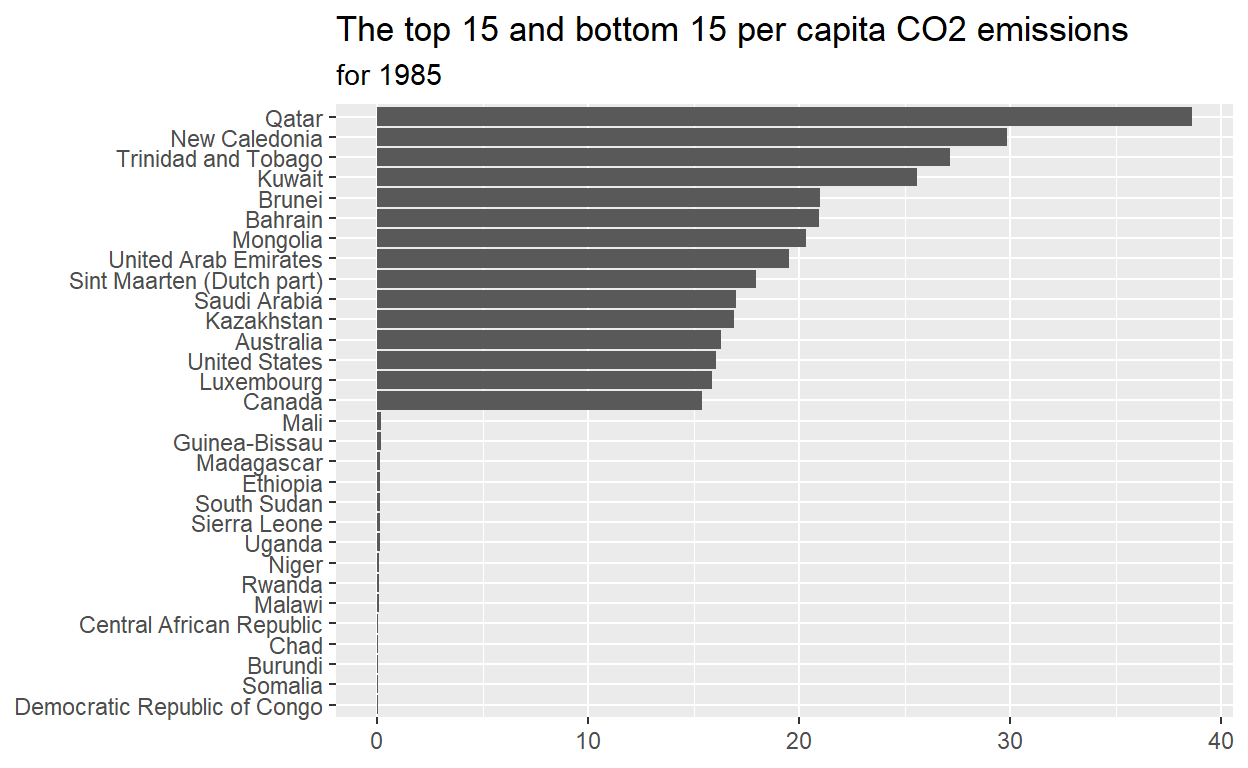
- Which 15 countries have the highest ‘per_capita_co2_emissions’?
start with ‘emissions_1985’ THEN use ‘slice_max’ to extract the 15 rows with the ‘per_capita_co2_emissions’ assign the output to ‘max_15_emitters’
max_15_emitters <- emissions_1985 %>%
slice_max(per_capita_co2_emissions, n = 15)
- Which 15 countries have the lowest ‘per_capita_co2_emissions’?
start with ‘emissions_1985’ THEN use ‘slice_min’ to extract the 15 rows with the ‘per_capita_co2_emissions’ assign the output to ‘min_15_emitters’
min_15_emitters <- emissions_1985 %>%
slice_min(per_capita_co2_emissions, n = 15)
- Use ‘bind_rows’ to bind together the ‘max_15_emitters’ and ‘min_15_emitters’ assign the output to ‘max_min_15’
max_min_15 <- bind_rows(max_15_emitters, min_15_emitters)
- Export ‘max_min_15’ to 3 file formats
max_min_15 %>% write_csv("max_min_15.csv") #comma-separated values
max_min_15 %>% write_tsv("max_min_15.tsv") #tab separated
max_min_15 %>% write_delim("max_min_15.psv", delim = "|") #pipe-separated
- Read the 3 file formats into R
max_min_15_csv <- read_csv("max_min_15.csv") #comma-separated values
max_min_15_tsv <- read_tsv("max_min_15.tsv") #tab separated
max_min_15_psv <- read_delim("max_min_15.psv", delim = "|") #pipe-separated
- Use ‘setdiff’ to check for any differences among ‘max_min_15_csv’, ‘max_min_15_tsv’ and ‘max_min_15_psv’
setdiff(max_min_15_csv, max_min_15_tsv, max_min_15_psv)
# A tibble: 0 x 3
# ... with 3 variables: country <chr>, code <chr>,
# per_capita_co2_emissions <dbl>Are there any differences?
- Reorder ‘country’ in ‘max_min_15’ for plotting and assign to man_min_15_plot_data
start with ‘emissions_1985’ THEN use ‘mutate’ to reorder ‘country’ according to ‘per_capital_co2_emissions’
max_min_plot_data <- max_min_15 %>%
mutate(country = reorder(country, per_capita_co2_emissions))
- Plot ‘max_min_15_plot_data’
ggplot(data = max_min_plot_data, mapping = aes(x = per_capita_co2_emissions, y = country)) + geom_col() + labs(title = 'The top 15 and bottom 15 per capita CO2 emissions', subtitle = "for 1985", x = NULL, y = NULL)
- Save the plot directory with this post
ggsave(filename = "preview.png",
path = here("_posts", "2021-02-26-reading-and-writing-data"))
- Add preview.png to yanl chuck at the top of this file
preview:preview.png